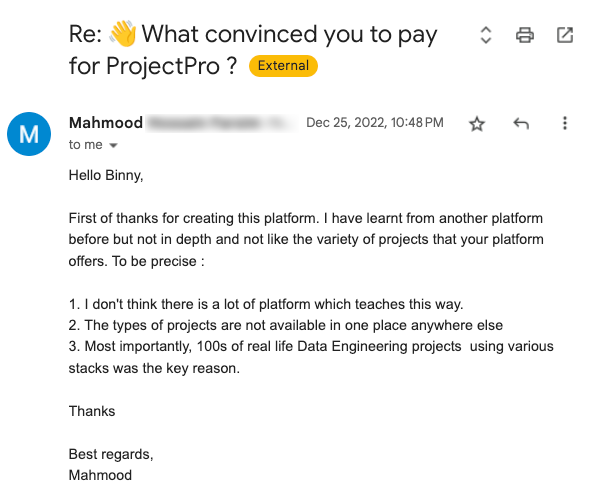
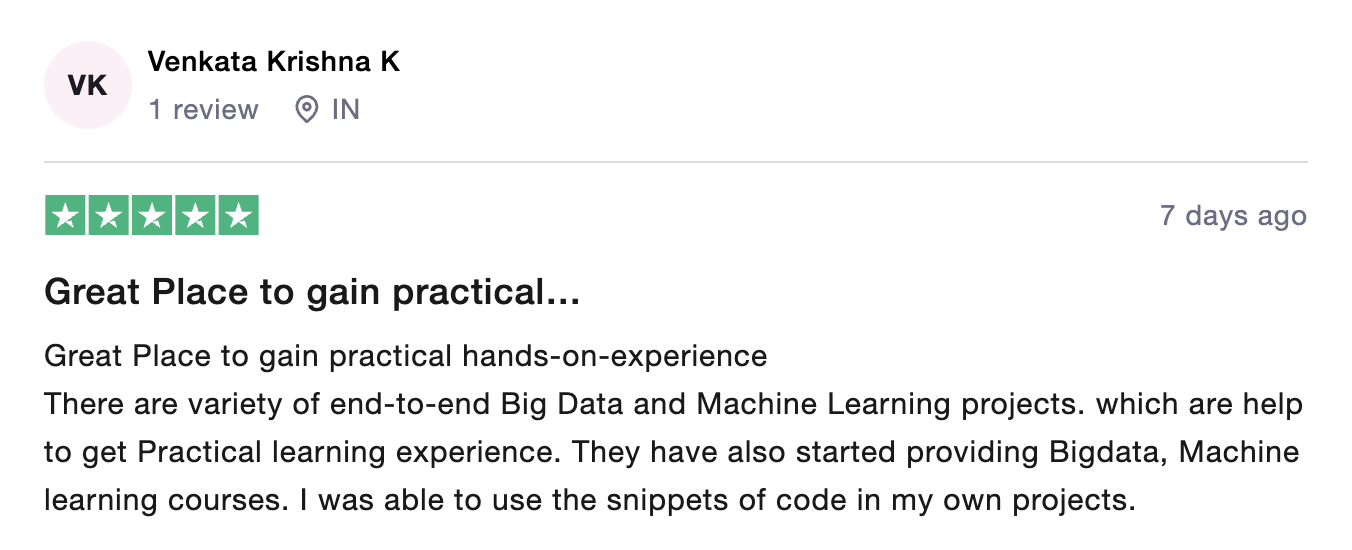
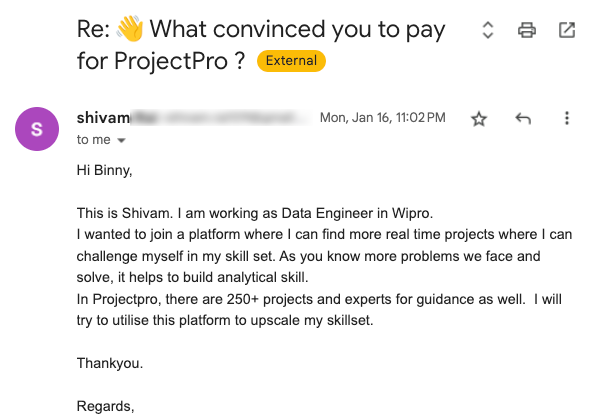
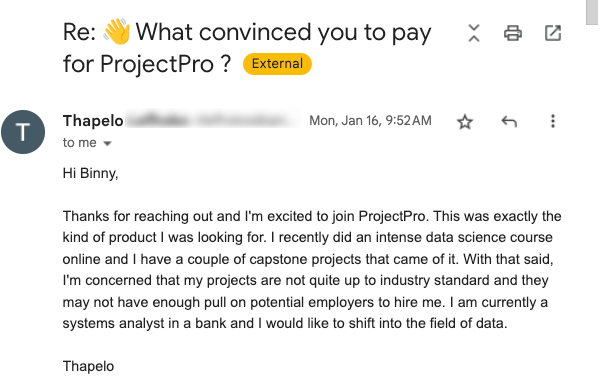
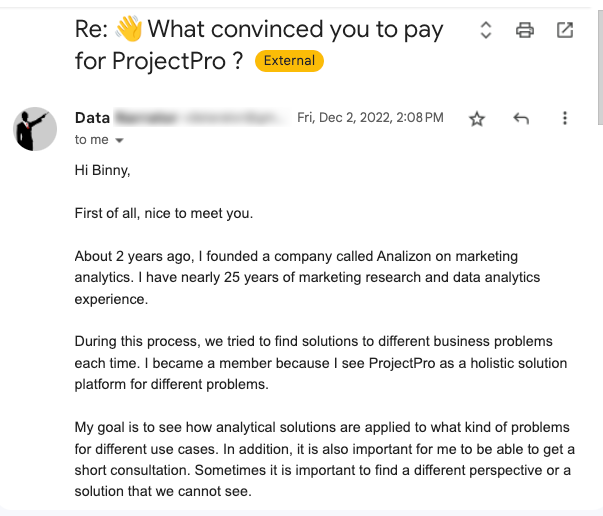
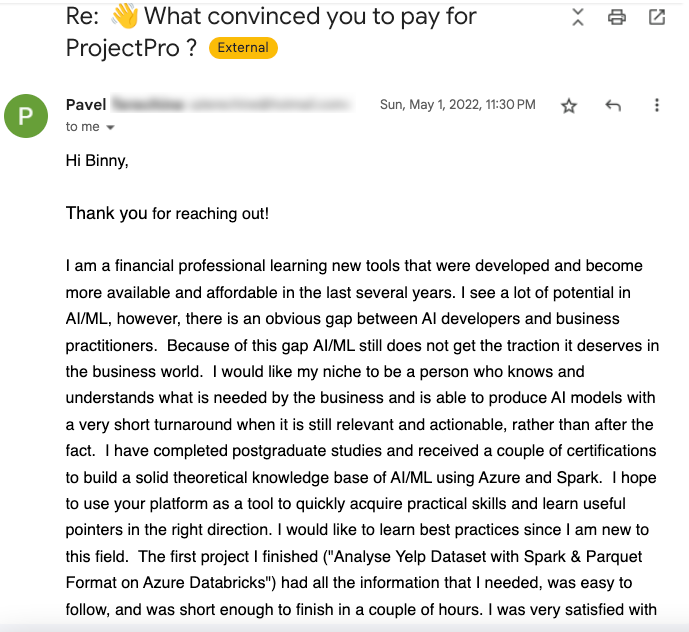
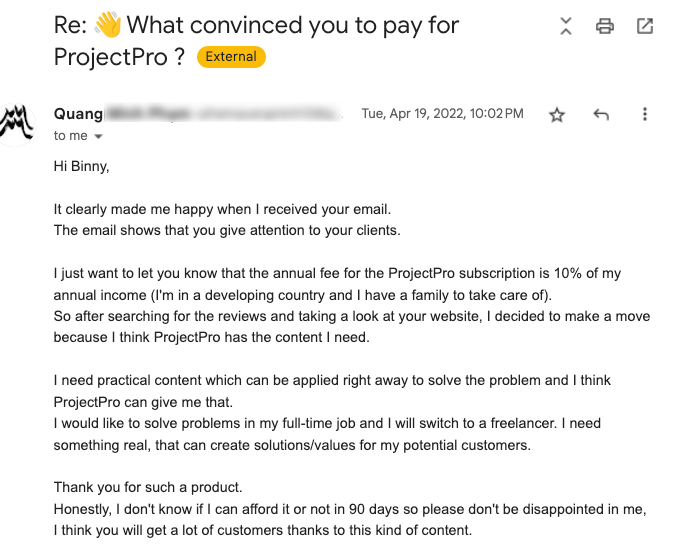
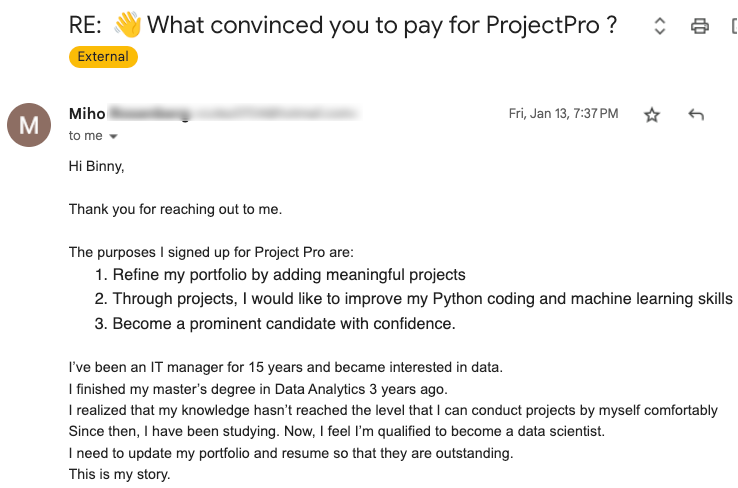
Global Data Community Lead | Lead Data Scientist, Thoughtworks

Dev Advocate, Pinecone and Freelance ML

Data Scientist, Inmobi

Senior Data Engineer, Publicis Sapient
In this Personalized Medicine Machine Learning Project you will learn to classify genetic mutations on the basis of medical literature into 9 classes.
Get started today
Request for free demo with us.
Schedule 60-minute live interactive 1-to-1 video sessions with experts.
Unlimited number of sessions with no extra charges. Yes, unlimited!
Give us 72 hours prior notice with a problem statement so we can match you to the right expert.
Schedule recurring sessions, once a week or bi-weekly, or monthly.
If you find a favorite expert, schedule all future sessions with them.
Source: ![]()
Source: ![]()
Source: ![]()
Source: ![]()
Source: ![]()
Source: ![]()
Source: ![]()
Source: ![]()
Source: ![]()
Source: ![]()
Source: ![]()
Source: ![]()
Source: ![]()
250+ end-to-end project solutions
Each project solves a real business problem from start to finish. These projects cover the domains of Data Science, Machine Learning, Data Engineering, Big Data and Cloud.
15 new projects added every month
New projects every month to help you stay updated in the latest tools and tactics.
500,000 lines of code
Each project comes with verified and tested solutions including code, queries, configuration files, and scripts. Download and reuse them.
600+ hours of videos
Each project solves a real business problem from start to finish. These projects cover the domains of Data Science, Machine Learning, Data Engineering, Big Data and Cloud.
Cloud Lab Workspace
New projects every month to help you stay updated in the latest tools and tactics.
Unlimited 1:1 sessions
Each project comes with verified and tested solutions including code, queries, configuration files, and scripts. Download and reuse them.
Technical Support
Chat with our technical experts to solve any issues you face while building your projects.
7 Days risk-free trial
We offer an unconditional 7-day money-back guarantee. Use the product for 7 days and if you don't like it we will make a 100% full refund. No terms or conditions.
Payment Options
0% interest monthly payment schemes available for all countries.
Overview
Personalized-Medicine-Redefining-Cancer-Treatment is a problem of classifying the given genetic mutations based on the literature available in the medical domain into one of the given nine classes. A lot has been said during the past several years about precision medicine and, more concretely, how genetic testing will disrupt the way diseases like cancer are treated. But this is only partially happening due to the huge amount of manual work still required. In this project, we will try to take personalized medicine to its full potential. Once sequenced, a cancer tumor can have thousands of genetic mutations. But the challenge is distinguishing the mutations contributing to tumor growth (drivers) from the neutral mutations (passengers).

Currently, this interpretation of genetic mutations is being made manually. This is a very time-consuming task where a clinical pathologist has to manually review and classify every single genetic mutation based on evidence from text-based clinical literature. The machine learning solution to this problem will help to speed up this time-consuming procedure and produce results with significant accuracy.
In this project, we create features from medical literature data and develop a machine-learning algorithm that automatically classifies genetic variations using this knowledge base as a baseline.
We will experiment with different models such as Logistic Regression, Random Forest, KNN, and Naive Bayes to find the best-fitting model for the problem statement.
Aim
To classify genetic mutations based on medical literature into the given nine classes.
Data Description
The dataset is divided into variants and text for training and test datasets, which includes the following features:
Fields are ID (the id of the row used to link the mutation to the clinical evidence)
Gene (the gene where this genetic mutation is located)
Variation (the aminoacid change for these mutations)
Class (1-9 the class this genetic mutation has been classified on)
Training_text (ID, Text), Text (the clinical evidence used to classify the genetic mutation)
Tech Stack
Language: Python
Libraries: pandas, numpy, pretty_confusion_matrix, matplotlib, sklearn, pymongo[srv]
Approach
Data Reading
Data Analysis
Class
Gene
Variation
Text Preprocessing
Splitting Data, Evaluation, and Features Extraction
Model Building
Logistic Regression
Random Forest
KNN
Naive Bayes
Hyperparameter Tuning
Logistic Regression
Model Evaluation
Confusion matrix
Log Loss
Recommended
Projects
Best MLOps Certifications To Boost Your Career In 2024
Chart your course to success with our ultimate MLOps certification guide. Explore the best options and pave the way for a thriving MLOps career. | ProjectPro
A comprehensive guide on LLama2 architecture, applications, fine-tuning, tokenization, and implementation in Python.
Adaboost Algorithm Explained in Depth
Exploring the AdaBoost Algorithm Applications, Working and Projects in Python.| ProjectPro
Get a free demo